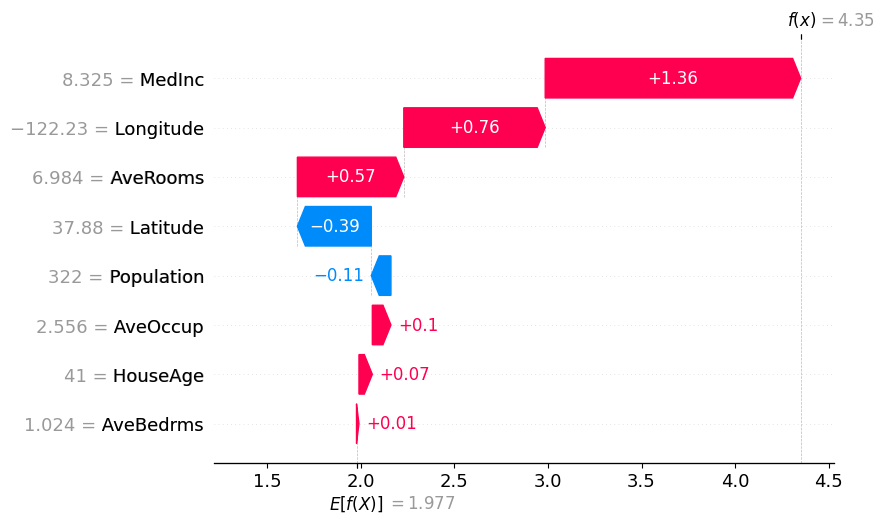
Simple California Demo
This notebook shows how to use build a hierarchical clustering of the input features and use it to explain a single instance. This is a good way to explain a single instance when the number of input features is large. When given a balanced partition tree PartitionExplainer has \(O(M^2)\) runtime, where \(M\) is the number of input features. This is much better than the \(O(2^M)\) runtime of KernelExplainer.
[1]:
import sys
import matplotlib.pyplot as plt
import numpy as np
import scipy as sp
import scipy.cluster
from xgboost import XGBRegressor
import shap
seed = 2023
np.random.seed(seed)
Train a model
[2]:
X, y = shap.datasets.california()
model = XGBRegressor(n_estimators=100, subsample=0.3)
model.fit(X, y)
instance = X[0:1]
references = X[1:100]
is_sparse is deprecated and will be removed in a future version. Check `isinstance(dtype, pd.SparseDtype)` instead.
is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
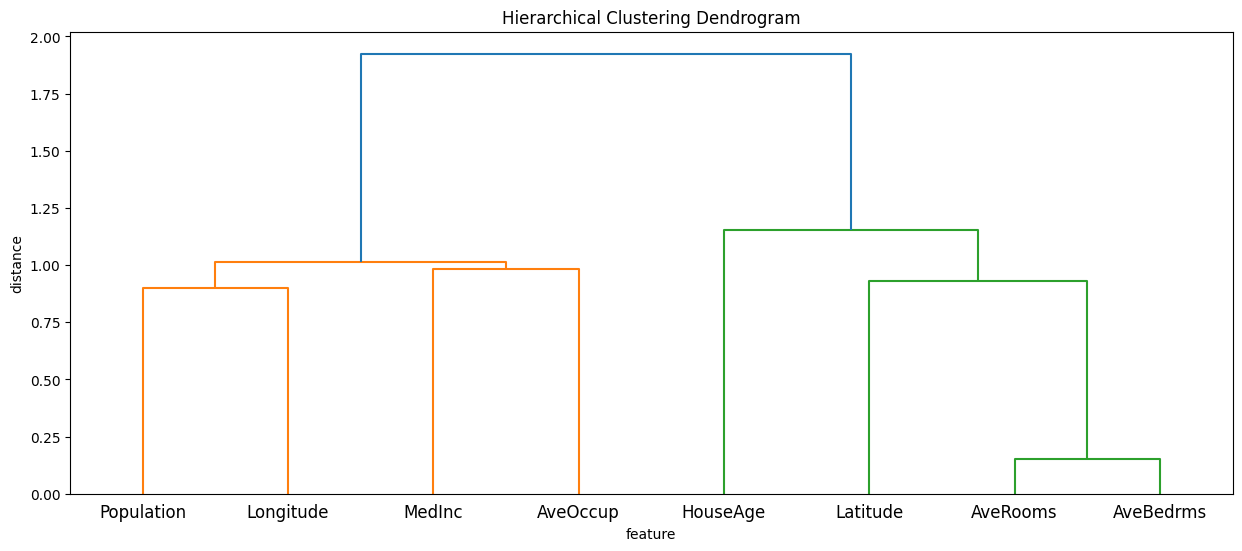
Compute a hierarchical clustering of the input features
[3]:
partition_tree = shap.utils.partition_tree(X)
plt.figure(figsize=(15, 6))
sp.cluster.hierarchy.dendrogram(partition_tree, labels=X.columns)
plt.title("Hierarchical Clustering Dendrogram")
plt.xlabel("feature")
plt.ylabel("distance")
plt.show()
Explain the instance
[4]:
# build a masker from partition tree
masker = shap.maskers.Partition(X, clustering=partition_tree)
# build explainer objects
raw_explainer = shap.PartitionExplainer(model.predict, X)
masker_explainer = shap.PartitionExplainer(model.predict, masker)
# compute SHAP values
raw_shap_values = raw_explainer(instance)
masker_shap_values = masker_explainer(instance)
[5]:
# comparison the masker and the original data sizes
print(f"X size: {sys.getsizeof(X)/1024:.2f} kB")
print(f"masker size: {sys.getsizeof(masker)} B")
X size: 1290.16 kB
masker size: 56 B
Compare to Tree SHAP
[6]:
tree_explainer = shap.TreeExplainer(model, X)
tree_shap_values = tree_explainer(instance)
plt.figure(figsize=(15, 6))
plt.plot(tree_shap_values[0].values, label="Tree SHAP")
plt.plot(masker_shap_values[0].values, "g--", label="Partition SHAP")
plt.plot(raw_shap_values[0].values, "r--", label="Raw SHAP")
plt.legend()
plt.show()
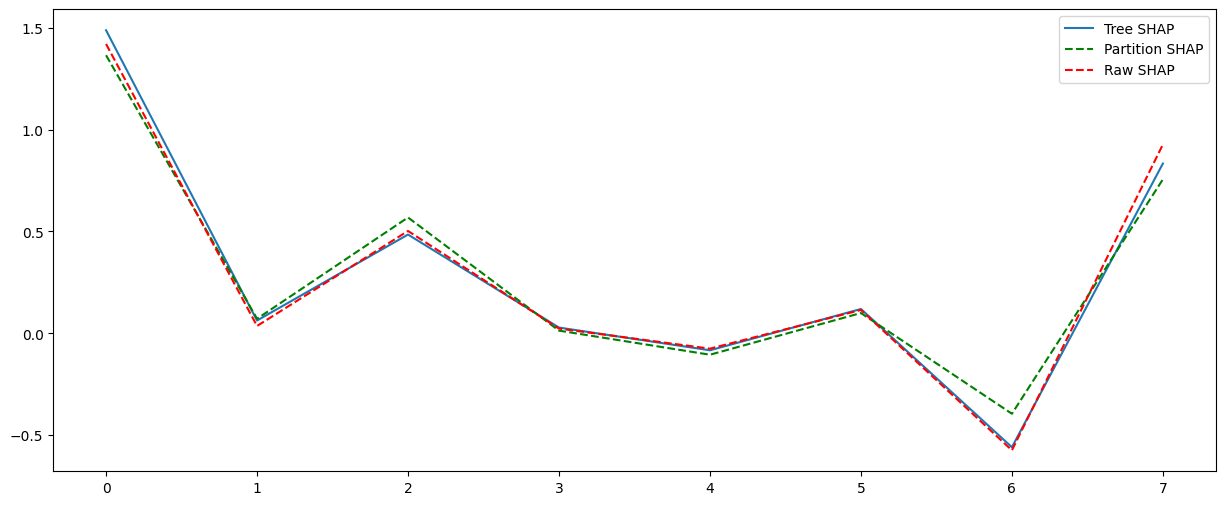
Partition SHAP values using a partition tree are nice estimation of SHAP values. The partition tree is a good way to reduce the number of input features and speed up the computation.
Plots to explain the instance
[7]:
shap.plots.waterfall(masker_shap_values[0])